- Joined
- May 30, 2004
- Location
- Folding@Home
.

The epidermal growth factor receptor (EGFR) kinase catalytic domain bound to a small-molecule experimental inhibitor. PDB accession code: 2HZ0. Image by Daniel L. Parton.
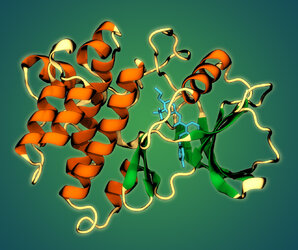
The epidermal growth factor receptor (EGFR) kinase catalytic domain bound to a small-molecule experimental inhibitor. PDB accession code: 2HZ0. Image by Daniel L. Parton.
Fighting cancer on Folding@Home: EGFR
January 14, 2014 by john Chodera ·
[Guest post by Daniel L. Parton of the Chodera Lab, Memorial Sloan-Kettering Cancer Center.]
We’re about to roll out our first major F@h project on the new work server at Memorial Sloan-Kettering Cancer Center (MSKCC). Our chosen target is a protein known as the epidermal growth factor receptor (EGFR), which is a member of the kinase family of human proteins. Kinases are key components in cellular signaling pathways, which control and coordinate all cellular activities. When kinases are activated, they pass messages to other proteins by attaching small phosphate molecules to specific parts of their structure. For proper regulation of signaling pathways, the cell must also be able to temporarily inactivate kinases. However, mutations in kinase genes may cause them to become constantly active, resulting in overactivation of the associated signaling pathways. In some cases, this causes the cell to begin dividing and growing uncontrollably, and this chain of events is the cause of many types of cancer.
EGFR mutations are specifically associated with certain types of cancer of the lung, breast, colon, rectum, and brain. Although a small number of anti-EGFR drugs are available (for example, gefitinib, erlotinib, and cetuximab), the development of resistance mutations during treatment is a huge barrier to their effective use.
The Chodera Lab is working to understand a number of factors which have historically hindered the development of effective kinase drug treatments. Firstly, we are interested in what makes a drug molecule selective for one kinase over another (since there are approximately 500 types of kinase in a human cell with nearly identical active sites). Secondly, we want to understand the physical mechanism by which resistance mutations develop to diminish the therapeutic effect of a drug. Eventually, we also hope to be able to use genetic sequencing data from individual cancer patients to predict which particular drug treatment might be optimal given a particular patient’s tumor mutations. There are many theoretical and algorithmic hurdles that we will need to overcome on the way, but these F@h simulations of EGFR will be a first step towards these goals.
Thanks again to all the F@h donors for contributing their computer time to enable this research!
–
The team of researchers studying EGFR includes Daniel L. Parton, Patrick M. Grinaway, Kyle A. Beauchamp, and Sonya M. Hanson in the Chodera Lab at MSKCC. Special thanks to Richard Knospler in the MSKCC Open Systems group for all of his hardware and software help in getting our servers up and running!
 for this project
for this project